Gene and protein expression of epithelial to mesenchymal transition for intestinal and anal fistula: a systematic review
Article information
Abstract
Purpose
Intestinal fibrosis is a common complication of inflammatory bowel diseases. However, the possible involvement of epithelial-mesenchymal transition (EMT) has been scarcely investigated. This systematic review aims to search through research papers that are focusing on messenger RNA (mRNA) and protein expression profile in EMT in fistula or in intestinal fibrosis.
Methods
Electronic exploration was performed until April 24, 2019 through PubMed, Ovid, Science Direct, and Scopus databases with the terms of “fistula” OR “intestinal fibrosis” AND “epithelial-mesenchymal transition”. Two independent reviewers scrutinized the suitability of the title and abstract before examining the full text that met the inclusion criteria. For each study, the sample types that were used, methods for analysis, and genes expressed were identified. The list of genes was further analyzed using DAVID (Database for Annotation, Visualization, and Integrated Discovery) and KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway.
Results
There were 896 citations found; however, only 3 studies fulfilled the requirements. Among the EMT-related genes, 5 were upregulated genes at mRNA level while 6 were at protein level. However, only 2 downregulated genes were found at each mRNA and protein level. Of the 4 inflammation-related genes found, 3 genes were upregulated at mRNA level and 1 at protein level. These genes were confirmed to be involved in the development of inflammatory induced fibrosis and fistula through EMT. Results from quantitative real-time polymerase chain reaction analysis were consistent with the process of EMT, confirmed by the western blot protein analysis.
Conclusion
Many significant genes which are involved in the process of EMT in fistula and intestinal fibrosis have been identified. With high-end technology many more genes could be identified. These genes will be good molecular targets in the development of biomarkers for precision drug targeting in the future treatment of intestinal fibrosis and fistula.
INTRODUCTION
Intestinal fibrosis is defined as disproportionate deposition of extracellular matrix (ECM) as a result of chronic inflammation and deficient intestinal wound healing with an underlying expansion of mesenchymal cells [1, 2]. This complication is commonly found in inflammatory bowel diseases (IBDs), whereby it affects more than 30% and almost 5% of patients with Crohn disease (CD) and ulcerative colitis, respectively [3–5]. Fibrosis and stricture formation frequently proceed to fistula formation, indicating that both processes may be linked or interdependent. Although most of the IBD patients exhibit inflammatory disease phenotype, one-third of them show evidence of stricture or penetrating intestinal complication at the time of diagnosis. However, the pathogenesis of fistula formation is still unknown [6]. To date there are no anti-fibrotic drugs available that can prevent or treat intestinal fibrosis, making surgery the sole therapeutic solution. Unfortunately, once the process starts the evolution of fibrosis and stricture formation cannot be prevented through the use of anti-inflammatory drugs, be it in IBD or other chronic inflammatoryassociated fibrotic conditions of various organs (liver, kidney, and lung) [7–11].
Intestinal fibrosis is the predominant factor for luminal stricture and obstruction which leads to dysfunction of the affected digestive tracts [12]. The fibrosis phenomenon is represented by disproportionate production and deposition of ECM components through the effector cell which is the activated myofibroblasts. However, the source of the myofibroblasts was previously poorly understood until novel studies suggest that a significant fragment of synthetically active myofibroblasts arises from the conversion of epithelial cells through the process of epithelial-mesenchymal transition (EMT) [13–15]. Generally, EMT is involved in both physiological and pathological processes which include embryogenesis, organ development, wound healing, tissue remodeling, tissue fibrosis, and cancer progression [6]. During the process of EMT, the epithelial cells lose their cell-cell contacts and their distinctive protein expression patterns such as epithelial cadherin (E-cadherin), catenins, and cytokeratins, and gain phenotypic characteristics of mesenchymal cells or fibroblast markers like fibroblast-specific protein-1, alpha smooth muscle actin (α-SMA) and vimentin [2, 16, 17]. This is partially due to the coordinated regulation of a unique set of transcription factors, such as Snail1, Slug, or Twist [6,18]. In this systematic review, the literatures on gene and protein expression involved in EMT for fistula or intestinal fibrosis are reviewed in order to establish the most prevalent gene/protein expression signature. By studying the gene/protein expression, we could then possibly provide new understandings into the regulation of genes in EMT in fistula or intestinal fibrosis. This study offers new knowledge in explaining the basis of identification of the changes toward the mechanism attributed by EMT in fistula or intestinal fibrosis.
METHODS
Data sources and search strategy
The search of contemporary articles was accomplished by using the PRISMA (Preferred Reporting Items for Systematic Reviews and Meta-Analyses) guidelines. Studies that fulfilled the inclusion criteria for the review were searched through PubMed, Ovid, Science Direct, and Scopus starting with the first original EMT article published until April 24, 2019. The following MeSH (Medical Subject Headings) terms of “fistula” OR “intestinal fibrosis” AND “epithelial-mesenchymal transition” were used.
Inclusion and exclusion criteria
Only English-language publications were incorporated for screening, while publication types without primary data, such as the letters to editors, editorials, case reports, scientific reports, conference proceedings, chapters in a book, and narrative reviews were excluded. Studies on EMT type 1 and 3, and studies not related to EMT, fistula, or intestinal fibrosis were also excluded.
The inclusion criteria were as follows: (1) the study on gene expression in EMT type 2 in intestinal fibrosis or fistula tissue versus normal tissue, (2) analyses on human tissue only, (3) any method of measuring and analyzing protein and/or messenger RNA (mRNA) expression, and (4) where total samples used in the study were mentioned. Articles were excluded based on the following: (1) use of anything but resected human fistula or intestinal fibrosis tissue, including primary cultures, cell lines, and transfected cells or induced EMT, (2) use of animal tissue, and (3) interventional study.
Identification of relevant studies and data extraction
All articles from searchers were downloaded into an EndNote library (EndNote ver. X4.0.1, Clarivate). Manuscripts retrieved from the database searches were screened by the titles, the abstracts, and the entire articles to select the relevant studies. The screenings were conducted by 2 reviewers. The duplicated papers were discarded by EndNote. Full-text articles references were obtained and examined by 2 reviewers to meet the preset inclusion criteria. A third author was consulted on the eligibility where there were disagreements between the reviewers. Records on reasons for elimination were kept.
From the studies included, the data were extracted were tabulated by 1 author. Each study includes in detail the author, year of study, study design, sample size, sampling type, and method for gene analysis, and the gene identified were tabulated. Data entry was also ensured for each study after completing data extraction.
Study quality and analysis
The research quality was assessed by discussing the reported details of analysis among the authors. The authors focused on the results and the reported list of gene expressions. To verify the validity of the eligible studies, reviewers worked independently and with adequate reliability. For each study, bias was excluded by adhering to the inclusion criteria. The genes listed were further analyzed with the DAVID (Database for Annotation, Visualization, and Integrated Discovery; https://david.ncifcrf.gov/home.jsp) [19]. Further analysis was performed to determine the cluster of genes that displayed significant functional annotation enrichment and those enriched annotations could be related to the EMT in fistula or in intestinal fibrosis, while the contribution of genes in the pathway was based on the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway [20].
RESULTS
The search strategies from 4 databases yielded 896 potentially relevant titles. Of these, 786 articles were eligible and had been retrieved for abstract review after duplicates were removed. Of the 786 studies that were reviewed initially, 61 were selected for full-text analysis. After comprehensive review of 61 full reports, only 3 articles fulfilled our inclusion and exclusion criteria hence selected for this systematic review. A PRISMA diagram describing the search results is shown in Fig. 1.
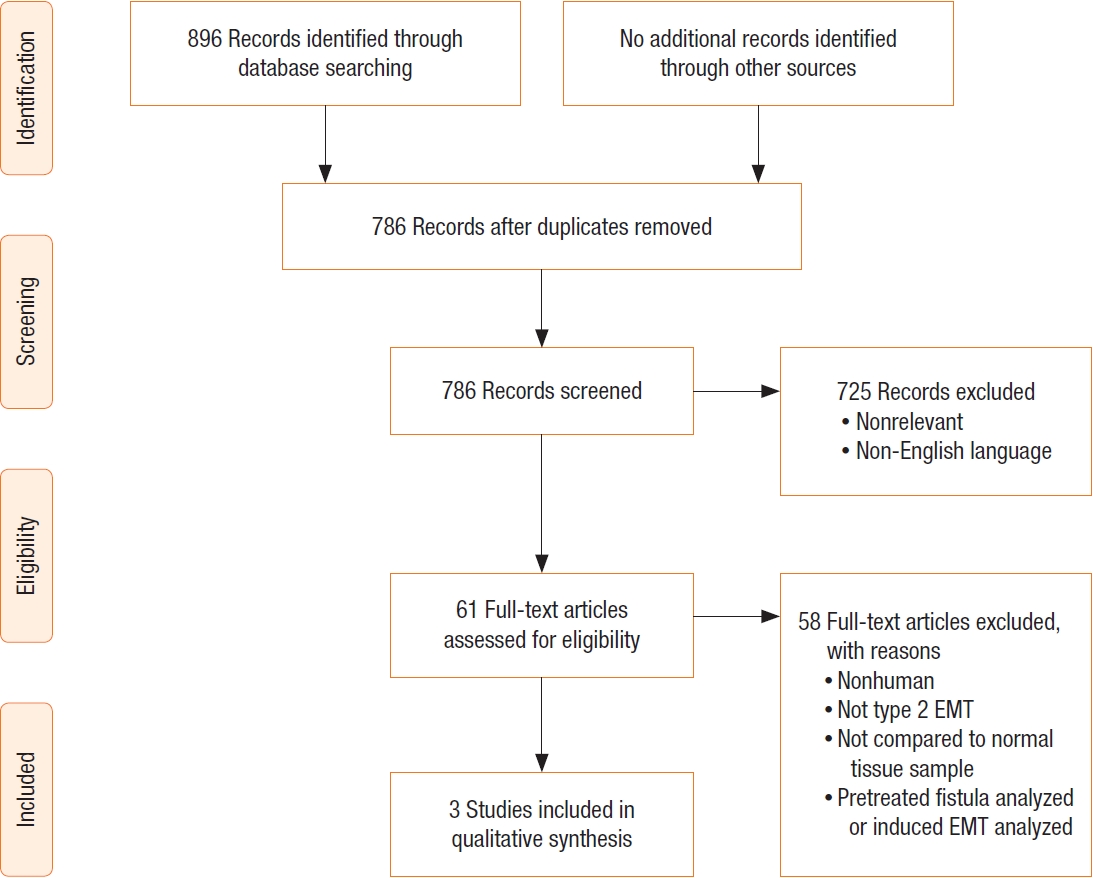
PRISMA (Preferred Reporting Items for Systematic Reviews and Meta-Analyses) flow diagram of search strategy and selection process. EMT, epithelial-mesenchymal transition.
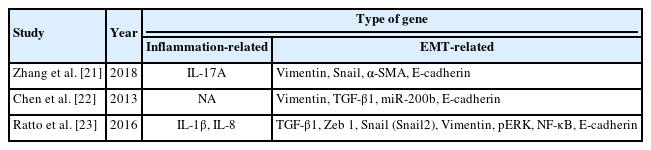
In total, 8 genes and 7 proteins were identified as being overexpressed and 2 genes and 2 proteins being under-expressed in EMT in cases of intestinal fibrosis or fistula compared to normal tissue. Tables 1 and 2 summarizes the features of these studies.
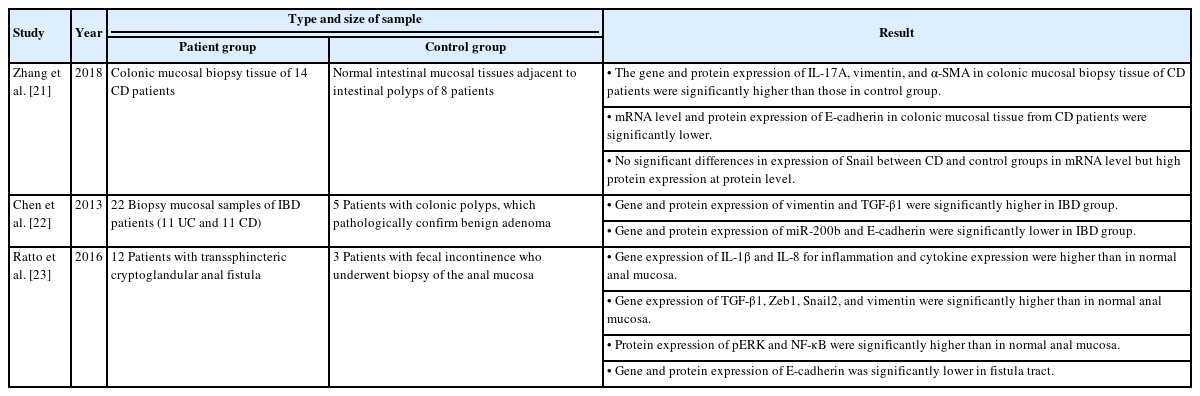
Two studies used IBD for the patient group whereby, mucosal tissue biopsies were taken from the colon. Zhang et al. [21] looked into the roles of interleukin (IL)-17A and EMT in the initiation and development of intestinal fibrosis while Chen et al. [22] studied the possible roles of microRNA-200b (miR-200b) in preserving intact intestinal epithelial cells (IECs) in IBDs. In another study, Ratto et al. [23] used samples from patients with cryptoglandular anal fistula to investigate the cytokine pattern that is potentially responsible for the inflammation, in assessing the evidence of EMT.
Detected genes during epithelial-mesenchymal transition process
A variety of biomarkers have been used to demonstrate the EMT process. Some are acquired and some are diminished. Gene expressions can be analyzed through many platforms such as microarray and gene sequencing as well as polymerase chain reaction (PCR). Quantitative real-time PCR (qRT-PCR) was used in all studies as a platform for analysis at the mRNA level. The protein expression was identified via western blotting technique in all studies.
In this review, we found that qRT-PCR was used to demonstrate the molecular characterization of the inflammation. Gene expression of IL-1β and IL-8 were found by Ratto et al. [23] to be higher in fistulas as compared to normal anal mucosa, with different levels of expression in the proximal and distal parts of the fistula tract. Meanwhile, Zhang et al. [21] found that the mRNA levels of IL-17A in colonic mucosal tissue of CD patients were significantly higher than those in the control groups.
The outcomes of the qRT-PCR analysis were consistent with a process of EMT. Among the genes analyzed, transforming growth factor beta 1 (TGF-β1) was overexpressed in studies by Chen et al. [22] and Ratto et al. [23] while vimentin was significantly higher in all 3 studies [21–23]. Zeb-1 and Snail were found highly expressed in fistula tract [23] while α-SMA was significantly higher in colonic mucosa of CD patients [21]. A reduction of E-cadherin was found in all of these 3 studies [21–23] and miR-200b was significantly lower in IBD groups [22].
Protein expression evaluated by western blot analysis confirmed the qRT-PCR findings. E-cadherin [21–23] and miR-200b [22] were expressed only in healthy anal mucosa, protein kinase RNA-like endoplasmic reticulum kinase (pERK) and nuclear factor kappa B (NF-κB) showed strong expression in fistula tract [23] while Snail and α-SMA were highly expressed in CD patients [21]. Vimentin [21, 22] and TGF-β1 [22] were overexpressed in all IBD patients (Fig. 2).

Summary of 3 different studies showing the upregulated (upward arrows) and downregulated (downward arrows) genes/proteins. Different color areas represent different datasets. The overlapping areas mean the common differentially expressed genes. IL, interleukin; α-SMA, smooth muscle alpha-actin; miR-200b, microRNA-200b; E-cadherin, epithelial cadherin; TGF-β1, transforming growth factor beta 1; pERK, protein kinase RNA-like endoplasmic reticulum kinase; NF-κB, nuclear factor kappa B.
Differentially expressed genes’ ontology analysis
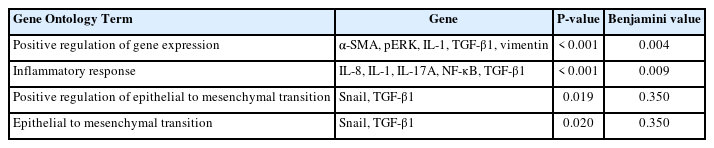
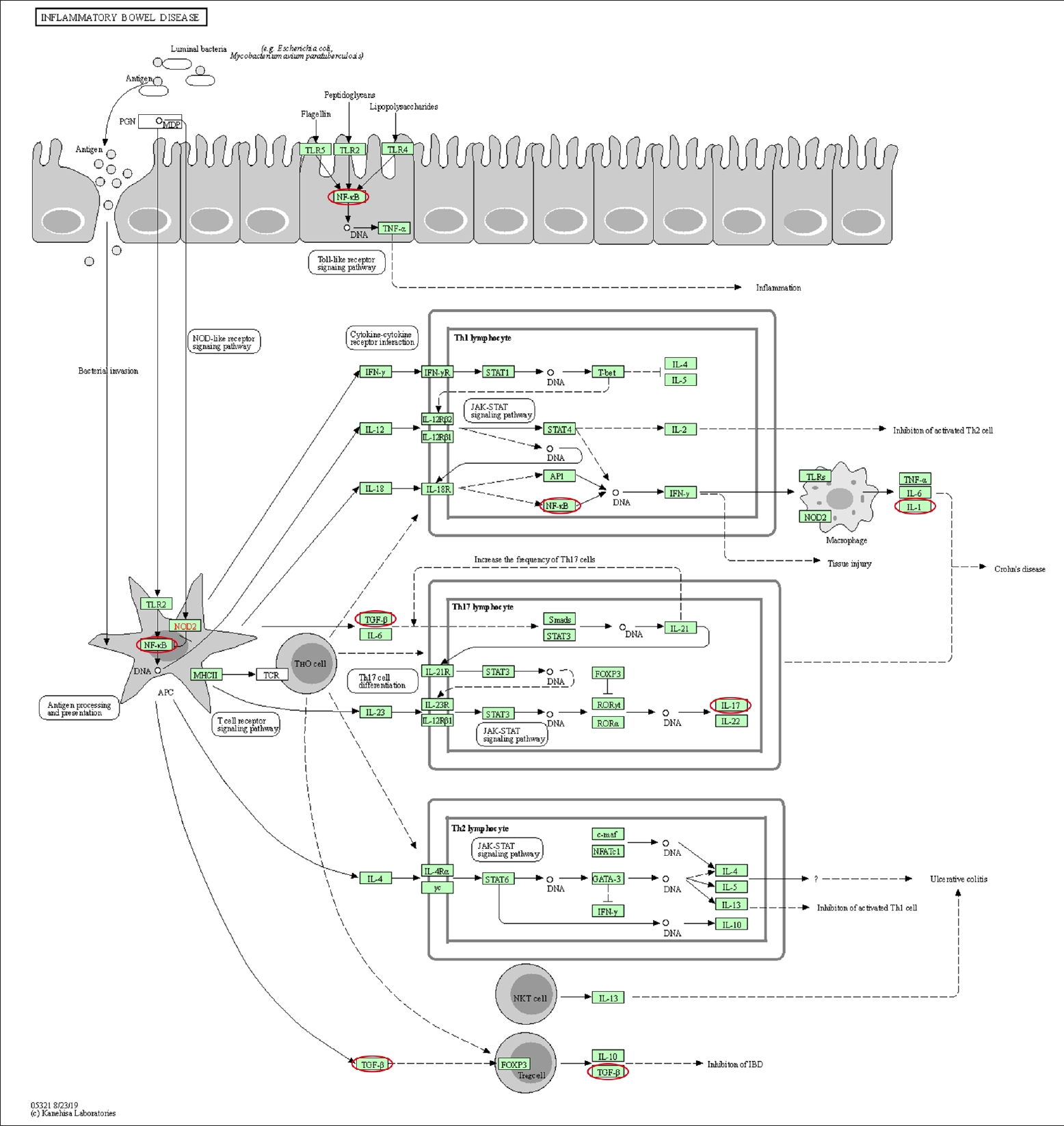
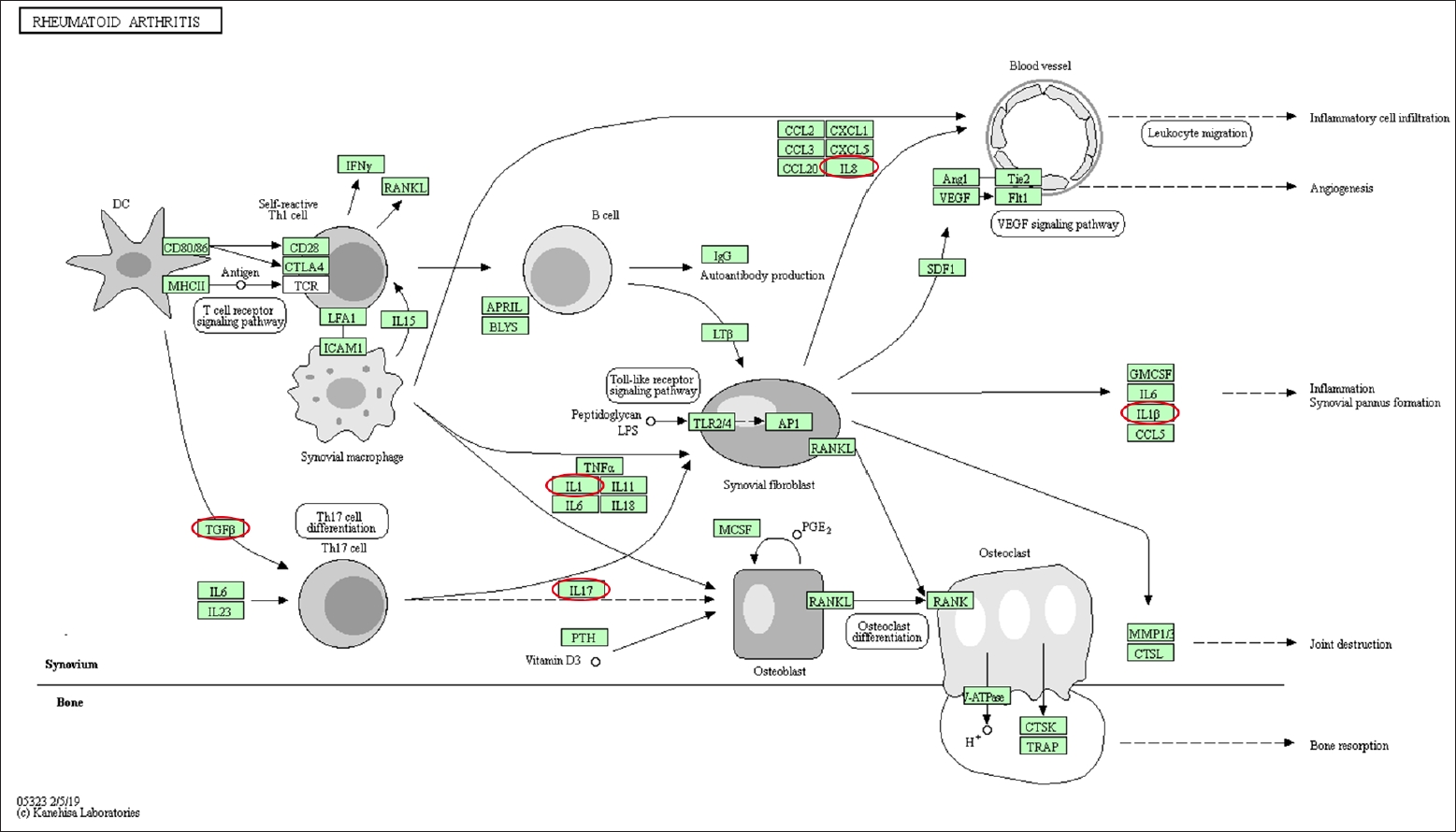
Differentially expressed genes and pathways enrichment were analyzed using DAVID [19]. From the Gene Ontology Term based on Expression Analysis Systematic Explorer (EASE) score, the positive regulation of gene expression exhibited the highest enrich term compared to the other terms which are related to this review study. A P-value of < 0.05 was used as cut-off to functionally characterize the genes, which were found to be differentially expressed in the different types of samples. The list of the genes involved is as shown in Table 3. Based on KEGG pathway analysis, there were 28 pathways involved for the selected genes. The top 3 pathways which were indirectly related to this review study are nonalcoholic fatty liver disease (NAFLD), IBD, and rheumatoid arthritis (Figs. 3–5). These 3 diseases are related to organ fibrosis and fistula diseases.
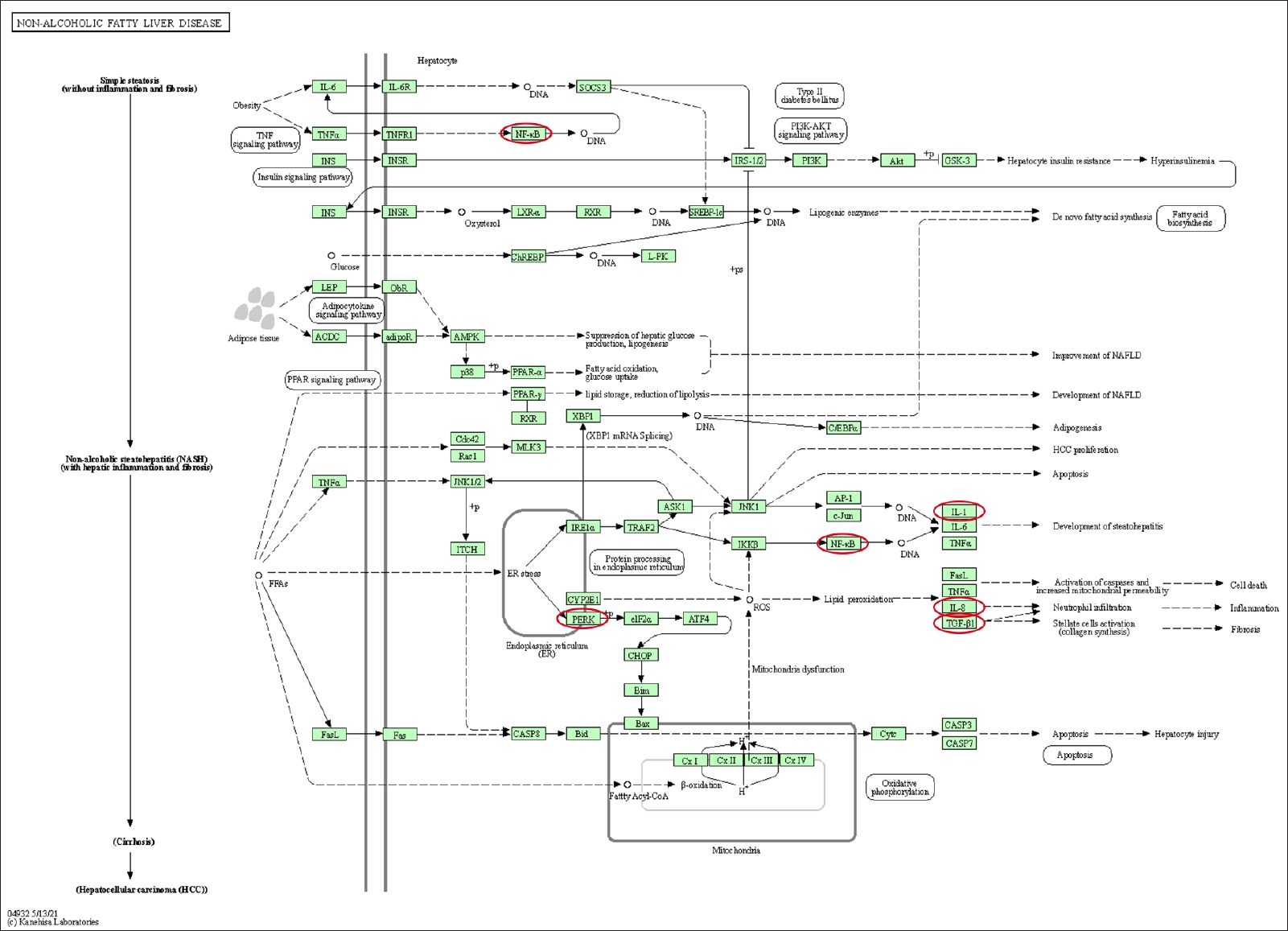
The KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway map of nonalcoholic fatty liver disease (map04932). The genes involved in this review study are shown in the red circles. Adapted from Kanehisa and Goto this review study are shown in the red circles. Adapted from Kanehisa and Goto [20] (reference no. 210879), according to the Creative Commons License.
DISCUSSION
Fibrosis is the most typical complication in IBD, which is considered a vital factor of intestinal stricture in CD patients. However, the mechanism of intestinal fibrosis is still a moot point among surgeons, hence becomes a big issue when deciding the appropriate treatment for the patients. Therefore, a recent study suggested that there are multiple mechanisms to reverse the fibrosis by molecular technique in IBD patients [24]. Fibroblasts and the epithelial cells are the main progenitor cells of activated myofibroblasts in intestinal fibrosis before they go through the EMT process [4, 5, 21, 25]. The complex process of EMT is characterized by losing the epithelial cell marker such as E-cadherin and gaining in the expression of mesenchymal features. The type 2 EMT processes which have been specifically observed are the loss of polarity of the epithelial cells, development of a specialized cytoskeleton, and invasive capacity by the intercellular adhesive molecules and membrane-bound transporter proteins [18, 26].
From this review, the results of the qRT-PCR analysis were consistent with a process of EMT and the evaluation of protein expression by western blot analysis confirmed the finding of qRT-PCR. In particular, TGF-β1, a key molecule in the EMT process and a potent inducer, was overexpressed in patients’ samples when compared with normal samples [22, 23]. High expressions of vimentin (a mesenchymal marker) in patients’ samples in all studies also demonstrated the consistency of the EMT process [21–23]. Vimentin is found in the mesenchymal cells of various types of tissue during their developmental stages to maintain the cells and tissue integrity [27].
The EMT process is also supported by the expression of E-cadherin. A reduction of E-cadherin expression was found in all the 3 studies [21–23]. E-cadherin (a typical marker of epithelial cells) was expressed only in healthy anal mucosa but absent in the proximal part of the fistula and was slightly detectable in the distal part [23]. The expression also decreased in the inflamed mucosa of IBD in comparison with the controls at both mRNA and protein level [21, 22]. Reduction of E-cadherin with a strong expression of pERK and NF-κB, the 2 intracellular factors which had been induced by TGF-β1 was also associated with the EMT process [23, 28]. A study by Chen et al. [22] found that miR-200b was significantly reduced in IBD and it correlated with the low expression of E-cadherin which was essential for the conservation of architecture and function of the IECs. The elevation of miR-200b expression in epithelium of IBD might oppose the loss of tight junctions and polarity in IECs [22].
Snail, a typical marker (transcriptional factors) in the EMT process showed expression in patients’ samples in only 2 studies which are done by Zhang et al. [21] and Ratto et al. [23]. Zhang et al. [21] found that there is no significant expression of Snail between the CD patients and control group at the mRNA level, but at the protein level, the expression shown was significantly higher than those in control groups. On the contrary, the expression of Snail was increased in patients who have fistula compared with healthy anal mucosa sample. However, there was no analysis for Snail done at the protein level [23]. Strong expression of Snail and lack in expression of E-cadherin may help to recognize the manifestation of EMT. Both of these genes contain enhancer box (E-box) sequences in their target gene promoter. However, as Snail is a specific transcriptional factor in EMT, its E-box sequence can recognize and inhibit the target gene expression which later decreases the expression of E-cadherin [29]. The discrepancy between the mRNA and protein levels in the study of Zhang et al. [21] may be due to various factors as small sample size or sample degradation.
Zeb1, another of the master regulators of EMT transcriptional program can also bind to the E-cadherin promoter and repress its transcription, thus leads to the low expression of E-cadherin. The low expression of E-cadherin with the high expression of Zeb1 in Ratto et al. [23] may help to show the existence of EMT. They found that Zeb1 was highly expressed while E-cadherin showed low expression in the fistula tract compared with normal anal mucosa at mRNA level. Meanwhile, α-SMA that has been recognized as a vital marker for differentiation and activation of myofibroblasts has been detected as an upregulated gene in Zhang et al. [21], both at mRNA and protein level in patients’ samples. The expression of α-SMA is one of the essential indicators for fibrosis. Fibrosis will develop when there is a huge number of aggregated and activated myofibroblasts which then produce and secrete a large number of ECM [4, 30].
In type 2 EMT, the process begins when inflammation occurs but will stop once the inflammation subsides, especially so during wound healing and tissue regeneration [31, 32]. However, during organ fibrosis, the EMT process will continue to respond to persistent inflammation, resulting in tissue destruction [31]. In this review, the molecular characterization of the inflammation was detected by the high expression of IL-1β and IL-8 in fistulas samples [23] and strong expression of IL-17A in CD patients [21]. IL-17A is related to the intestinal fibrosis development [33] which was confirmed by Zhang et al. [21]. More recent studies indicated that IL-17A might induce EMT on epithelial cells to indirectly promote fibrosis in multiple organs [34–38]. The abnormality in IL-17A expression can be a diagnostic marker for precursor and development of acute and chronic diseases.
Based on the KEGG pathway analysis, IL-1, IL-17, NF-κB, and TGF-β1 were involved in IBD pathway. Meanwhile the NF-κB, IL-1, IL-8, TGF-β1, and pERK were involved in the NAFLD. The KEGG pathway of rheumatoid arthritis involved the following genes: IL-1β, IL-8, TGF-β1, and IL-8 [20]. These pathways involved both the EMT-related genes as well as the inflammation-related genes. The coexistence of both types of genes on these pathways demonstrated that inflammation and EMT are interrelated.
Limited numbers of studies retrieved for this review had been primarily attributed by the strict preset criteria that had to be fulfilled namely, observational study without treatment or cell or tissue induction. This is to ensure that the EMT process occurred in vitro without being triggered by other factors. Hence, the studies that involved the in vivo EMT process were excluded. Theoretically, the EMT process occurred are due to continuous response to the inflammatory process which later leads to organ fibrosis. One other limitation is the small-sized samples in all the 3 studies which showed statistically nonsignificant results. The identification of control sample was another important and challenging issue as there was no standardized normal control tissue among this type of study, hence limiting the validity of the results. As qRT-PCR is the only method used, the list of genes is limited to the common genes involved. Microarray or next-generation sequencing analysis might be helpful in the detection of other genes that play a role in the EMT process in intestinal fibrosis.
Through this systematic review study, TGF-β1, Zeb1, Snail, vimentin, α-SMA, IL-1, IL-8, IL-8, pERK, and NF-κB had been identified as the genes being overexpressed genes while E-cadherin and miR-200b genes were under-expressed during the process of EMT in intestinal fibrosis. However, there is a need for a more comprehensive study on these genes and their protein expressions. These investigations will provide new insight into gene regulation and widen the basis of identification of the changes in the mechanisms of EMT in fistula or intestinal fibrosis. To date, surgery is the effective treatment for intestinal fibrosis and fistula in ano with the possibility of recurrence. The study on EMT-related genes can provide more insight into the pathophysiological course of the disease. A targeted adjunctive medical therapy can be made possible to reduce the need for surgical intervention as well as recurrence.
Notes
CONFLICT OF INTEREST
No potential conflict of interest relevant to this article was reported.
FUNDING
This research was funded by the Fundamental Research Grant Scheme awarded by the Ministry of Higher Education of Malaysia (USIM/FRGS/FPSK/055002/50317).
AUTHOR CONTRIBUTIONS
Conceptualization: RFJ, NHO, NAH; Data curation: RFJ, NHO; Formal analysis: RFJ, NHO; Funding acquisition: RFJ, HAR, NAH; Methodology: NHO; Supervision: RFJ, HAR, NAH; Validation: RFJ, HAR, NAH; Writing–original draft: NHO; Writing–review & editing: RFJ, HAR, NAH. All authors read and approved the final manuscript.